Adapting foundation models for medical image analysis re-
quires finetuning them on a considerable amount of data because of ex-
treme distribution shifts between natural (source) data used for pretrain-
ing and medical (target) data. However, collecting task-specific medical
data for such finetuning at a central location raises many privacy con-
cerns. Although Federated learning (FL) provides an effective means for
training on private decentralized data, communication costs in federat-
ing large foundation models can quickly become a significant bottleneck,
impacting the solution’s scalability. In this work, we address this prob-
lem of ‘efficient communication while ensuring effective learning in FL’ by
combining the strengths of Parameter-Efficient Fine-tuning (PEFT) with
FL. Specifically, we study plug-and-play Low-Rank Adapters (LoRA) in
a federated manner to adapt the Segment Anything Model (SAM) for
3D medical image segmentation. Unlike prior works that utilize LoRA
and finetune the entire decoder, we critically analyze the contribution of
each granular component of SAM on finetuning performance. Thus, we
identify specific layers to be federated that are very efficient in terms of
communication cost while producing on-par accuracy. Our experiments
show that retaining the parameters of the SAM model (including most of
the decoder) in their original state during adaptation is beneficial because
fine-tuning on small datasets tends to distort the inherent capabilities of
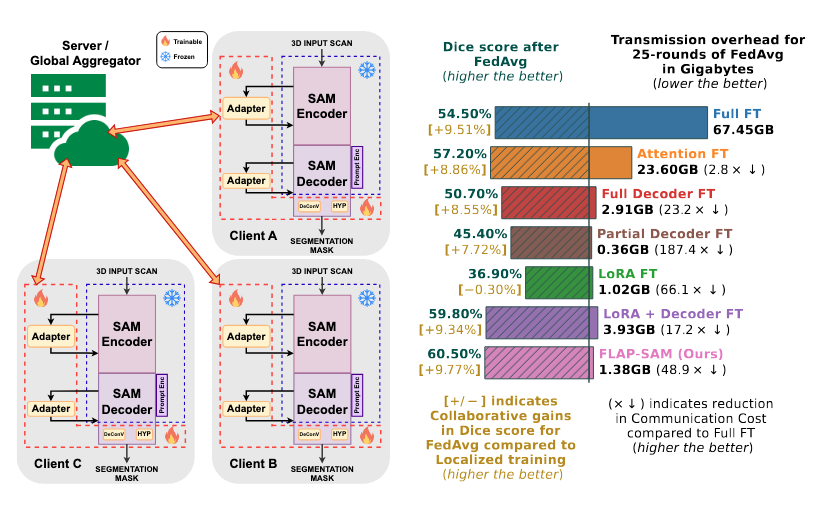
the underlying foundation model. On Fed-KiTS, our approach decreases
communication cost (∼48× ↓) compared to full fine-tuning while in-
creasing performance (∼6% ↑ Dice score) in 3D segmentation tasks. Our
approach performs similar to SAMed while achieving ∼2.8× reduction in
communication and parameters to be finetuned. We further validate our
approach with experiments on Fed-IXI and Prostate MRI datasets.
Paper Link: https://arxiv.org/pdf/2407.21739v1